'Deep-learning' computer system predicts genetic causes of diseases
5 January 2015
University of Toronto researchers have developed the first method for ranking genetic mutations based on how living cells read DNA, calculating how likely a mutation is to cause disease.
The multidisciplinary team from engineering, biology and medicine developed new computer techniques using massively parallel graphics processor units to analyse gene sequences and rate the likelihood of mutations causing diseases. So far they have discovered unexpected genetic determinants of autism, hereditary cancers and spinal muscular atrophy, a leading genetic cause of infant mortality. Their findings have been published in the journal Science.
“Over the past decade, a huge amount of effort has been invested into searching for mutations in the genome that cause disease, without a rational approach to understanding why they cause disease,” says team leader Professor Brendan Frey of the University of Toronto and also a senior fellow at the Canadian Institute for Advanced Research. “This is because scientists didn’t have the means to understand the text of the genome and how mutations in it can change the meaning of that text.”
The new approach
Certain sections of the genome, called exons, describe the proteins that are the building blocks of all living cells. What wasn’t appreciated until recently is that other sections, called introns, contain instructions for how to cut and paste exons together, determining which proteins will be produced. This ‘splicing’ process is a crucial step in the cell’s process of converting DNA into proteins, and its disruption is known to contribute to many diseases.
Most research into the genetic roots of disease has focused on mutations within exons, but increasingly scientists are finding that diseases can’t be explained by these mutations. Professor Frey’s team took a completely different approach, examining changes to text that provides instructions for splicing, most of which is in introns.
Frey’s team used a new technology called ‘deep learning’ to teach a computer system to scan a piece of DNA, read the genetic instructions that specify how to splice together sections that code for proteins, and determine which proteins will be produced. Unlike other machine-learning methods, deep learning can make sense of incredibly complex relationships, such as those found in living systems in biology and medicine.
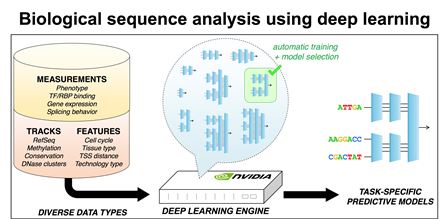
The
Deep Learning Engine: deep learning algorithms are used to train
computational models to predict phenotypes from biological
sequences, such as DNA, RNA or protein. Searches over a large number
of model architectures and training conditions are performed using
massively parallel GPU computing nodes.
"The success of our project relied crucially on using the latest deep learning methods to analyze the most advanced experimental biology data," said Frey, whose team included members from University of Toronto's Faculty of Applied Science & Engineering, Faculty of Medicine and the Terrence Donnelly Centre for Cellular and Biomolecular Research, as well as Microsoft Research and the Cold Spring Harbor Laboratory. "My collaborators and our graduate students and postdoctoral fellows are world-leading experts in these areas."
Once they had taught their system how to read the text of the genome, Frey’s team used it to search for mutations that cause splicing to go wrong. They found that their method correctly predicted 94% of the genetic culprits behind well-studied diseases such as spinal muscular atrophy and colorectal cancer, but more importantly, made accurate predictions for mutations that had never been seen before.
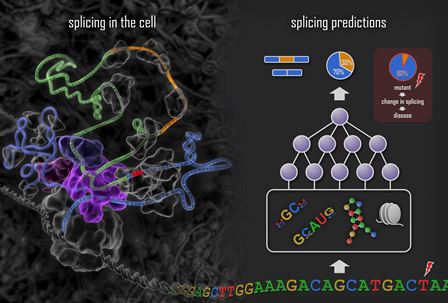
Deep learning techniques are used to train a
system that mimics the process of splicing in the cell (left panel).
Features such as motifs, RNA secondary structures and nucleosome
positions are computationally determined from the DNA sequence
(right panel), combinations of these features are combined to detect
complex patterns, and these are combined again to predict how
splicing will occur for the exon within the DNA sequence. The effect
of a DNA mutation is assessed by applying the system to the sequence
with and without the mutation and measuring the change in the
computed splicing level.
New genes connected to autism
The team then applied the system to analysing autism spectrum disorder, a condition with complex genetic underpinnings. “With autism there are only a few dozen genes definitely known to be involved and these account for a small proportion of individuals with this condition,” said Frey.
In collaboration with Dr. Stephen Scherer, senior scientist and director of The Centre for Applied Genomics at SickKids and the University of Toronto McLaughlin Centre, Frey’s team compared mutations discovered in the whole genome sequences of children with autism, but not in controls.
Following the traditional approach of studying protein-coding regions, they found no differences. However, when they used their deep learning system to rank mutations according to how much they change splicing, surprising patterns appeared.
“When we ranked mutations using our method, striking patterns emerged, revealing 39 novel genes having a potential role in autism susceptibility,” Frey said.
And autism is just the beginning — this mutation indexing method is ready to be applied to any number of diseases, and even non-disease traits that differ between individuals.
Dr. Juan Valcárcel Juárez, a researcher with the Center for Genomic Regulation in Barcelona, Spain, who was not involved in this research, says: “In a way it is like having a language translator: it allows you to understand another language, even if full command of that language will require that you also study the underlying grammar. The work provides important information for personalized medicine, clearly a key component of future therapies.”
Reference
Xiong HY et al. The human splicing code reveals new insights into the genetic determinants of disease. Science, December 18 2014. Abstract: www.sciencemag.org/lookup/doi/10.1126/science.1254806