ChipDX discovers genetic signature for early-stage colon cancer
21 Dec 2010
ChipDX LLC, a New York-based online molecular diagnostics and personalized medicine company, discovered and validated a genetic signature for early-stage colon cancer and is developing an online screening application to enable clinicians to more accurately identify risk of recurrence.
In the study, published in the December 2010 issue of the British Journal of Cancer, ChipDX demonstrates how the 163-gene signature stratifies colon cancer patients into high- and low-risk groups for recurrence with greater accuracy than current methods.
“We discovered a set of genes that were strongly associated with outcome, independent to current measurements of prognosis. By combining measurements of these genes, performed with Affymetrix GeneChip technology, and a robust predictive algorithm we were able to predict which individuals were at the greatest risk of recurrence within a five-year follow-up period,” said Ryan van Laar, PhD, ChipDX Founder and Chief Scientific Officer.
“This unique signature’s ability to generate a highly personalized assessment of recurrence risk may one day assist physicians in deciding whether individuals with early-stage colon cancer should receive chemotherapy in addition to surgery.”
Currently, ChipDX is making the algorithm available for research use only through an online gene expression analysis platform at www.ChipDX.com and is reviewing regulatory requirements and partners to market it as a test for future diagnostic use.
“Dr. van Laar’s discovery demonstrates the advancement in gene expression array applications and further points to the significance and relevance of this technology,” said Kevin Cannon, Vice President of Marketing, Gene Expression Applications, at Affymetrix.
“ChipDX’ online analysis platform embodies the Affymetrix goal of moving applications downstream from the basic research markets into clinical applications to vastly improve diagnosis and lead to better informed decisions for the most common cancers.”
Colon cancer, which causes 655,000 deaths worldwide each year, is the fourth most common form of cancer in the United States and the third leading cause of cancer-related death in the western world. Physicians currently use a method called clinical staging to measure the extent of disease spread at the time of diagnosis.
Patients who are diagnosed with early-stage tumours (1-2) generally do not receive chemotherapy; however, approximately 20% of stage 2 patients develop recurrence within five years. In the study, the ChipDX algorithm is shown to stratify early-stage colon cancer patients with greater accuracy than clinical staging.
“Ultimately, we hope our predictive gene signature will help doctors to identify these early-stage ‘high-risk’ patients and offer them more personalized treatment options based on a set of genes related to survival above and beyond traditional assessments of outcome,” said Dr. van Laar. “If treatment is tailored to the precise nature of a patient’s tumour, the life-saving potential is greater.”
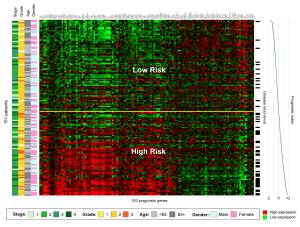 |
A gene expression 'heatmap' of the 163-probe signature in the training series is shown in Figure 2, in which genes (columns) are arranged by hierarchical clustering and patients (rows) are ordered according to their prognostic index. The relationship between gene expression and disease recurrence can be observed in the pattern of upregulation and downregulation formed by this arrangement, with higher expression of those genes on the left of the heatmap associated with poor prognosis and vice versa. An increasing frequency of recurrence events (indicated to the right of the heatmap) can be observed as the prognostic index increases from -2.0 to +2.0. (Graphic: Business Wire) |
ChipDX developed its prognostic algorithm by analyzing data from a 2009 study of 232 US-based colon cancer patients (Smith, et al., 2009)1. An independent validation series of 60 stage 2 and 3 Australian colon cancer patients was used to validate the discovery (Jorissen, et al., 2009)2. Both studies utilized Affymetrix GeneChip Human Genome U133 Plus 2.0 Array profiles of colon cancer patients. The method of gene expression data analysis performed was designed to identify genes significantly associated with recurrence independent of the patient’s age at diagnosis, tumour grade, or disease stage.
The ChipDX online platform performs multi-gene diagnostic analysis on whole-genome Affymetrix GeneChip data for research applications. An important component of the platform is the proprietary ChipDX Quality Module, developed by analyzing more than 3,000 GeneChip profiles of multiple tumor types generated in laboratories around the world.
This module ensures that each assay meets a high level of data integrity before a diagnostic analysis is performed. After this analysis, the data can be submitted to the newly created Colon Cancer Module, which uses the recently published 163-gene predictive algorithm to generate an individualized assessment of recurrence risk, in real-time. The ChipDX online analysis system is compatible with the Affymetrix GeneChip platform and currently available only for clinical research and assessment.
References
1. Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, Eschrich S, Kis C, Levy S, Washington MK, Heslin MJ, Coffey RJ, Yeatman TJ, Shyr Y, Beauchamp RD (2009) Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 138: 958–968.
2. Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, Kerr D, Aaltonen LA, Arango D, Kruhøffer M, Orntoft TF, Andersen CL, Gruidl M, Kamath VP, Eschrich S, Yeatman TJ, Sieber OM (2009) Metastasis-associated gene expression changes predict poor outcomes in patients with dukes stage B and C colorectal cancer. Clin Cancer Res 15: 7642–7651.